Clinical and inflammatory phenotyping by breathomics in chronic airway diseases irrespective of the diagnostic label
Aim: To capture clinical/inflammatory phenotypes in patients with chronic airway disease using an electronic nose (eNose) in a training and validation set.
Take home message: Exhaled breath analysis using eNose technology (the SpiroNose) can effectively identify clinical and inflammatory phenotypes in patients with chronic airway diseases, such as asthma and COPD, without relying on traditional diagnostic labels. The research demonstrated that eNose could detect systemic neutrophilia and eosinophilia in a dose-dependent manner, suggesting its potential as a non-invasive tool for rapid inflammatory phenotyping in chronic respiratory conditions.
Introduction
Asthma and chronic obstructive pulmonary disease (COPD) are recognized as complex, heterogeneous chronic airway diseases with diverse phenotypes, each having distinct inflammatory profiles and varying treatment responses. Despite improvements in care standards, the overlap between asthma and COPD still poses challenges in accurate diagnosis and individualized management.
Relying solely on traditional diagnostic labels like asthma and COPD may not fully capture a patient’s condition or inform tailored therapy. Instead, there is a shift towards a personalized approach focused on identifying “treatable traits.” This trend is supported by the rise of targeted therapies, which show improved outcomes when patients are selected based on specific inflammatory phenotypes, such as eosinophilia or neutrophilia.
Inflammatory phenotyping is typically done through blood or sputum analysis, but these tests are not practical for immediate results during patient visits. Molecular profiling of exhaled breath, if validated, offers a promising non-invasive alternative. While gas chromatography-mass spectrometry (GC-MS) can link VOCs in exhaled air to inflammatory profiles, it is not yet suitable for routine clinical use. In contrast, electronic nose (eNose) technology can analyze the full VOC profile through pattern recognition without identifying individual compounds. This method is quick, user-friendly, and has the potential to become a reliable point-of-care tool.
This study hypothesized that eNose technology could identify clinical and inflammatory profiles in chronic airway diseases independently of traditional asthma or COPD diagnoses. Using both unsupervised and supervised data analysis methods, the research aimed to uncover distinct patient phenotypes and link exhaled VOC patterns to inflammatory markers, followed by validation in a new cohort of asthma and COPD patients.
Methods
Study Population: BreathCloud is a multicenter cross-sectional study. This analysis includes the first 435 patients with asthma or COPD, split into a training set (n=321; asthma=206, COPD=115) and a validation set (n=114; asthma=72, COPD=42). Asthma and COPD patients were diagnosed based on international guidelines. Due to the non-invasive nature of exhaled breath analysis, ethics boards waived additional consent requirements. Clinical data, including lung function and blood counts, were collected as part of routine care.
Measurements: Breath analysis was performed using the SpiroNose®, which integrates eNose sensors with spirometry. Patients rinsed their mouths before two consecutive breath measurements, following a standardized protocol. Data were transmitted in real-time to the BreathBase® server for storage and analysis. Asthma and COPD patients were assessed using standardized questionnaires (ACQ for asthma, CCQ for COPD). Additional clinical data such as FEV1, atopy, FeNO, and blood counts were gathered. Exacerbations were defined as symptom worsening requiring oral corticosteroids or antibiotics.
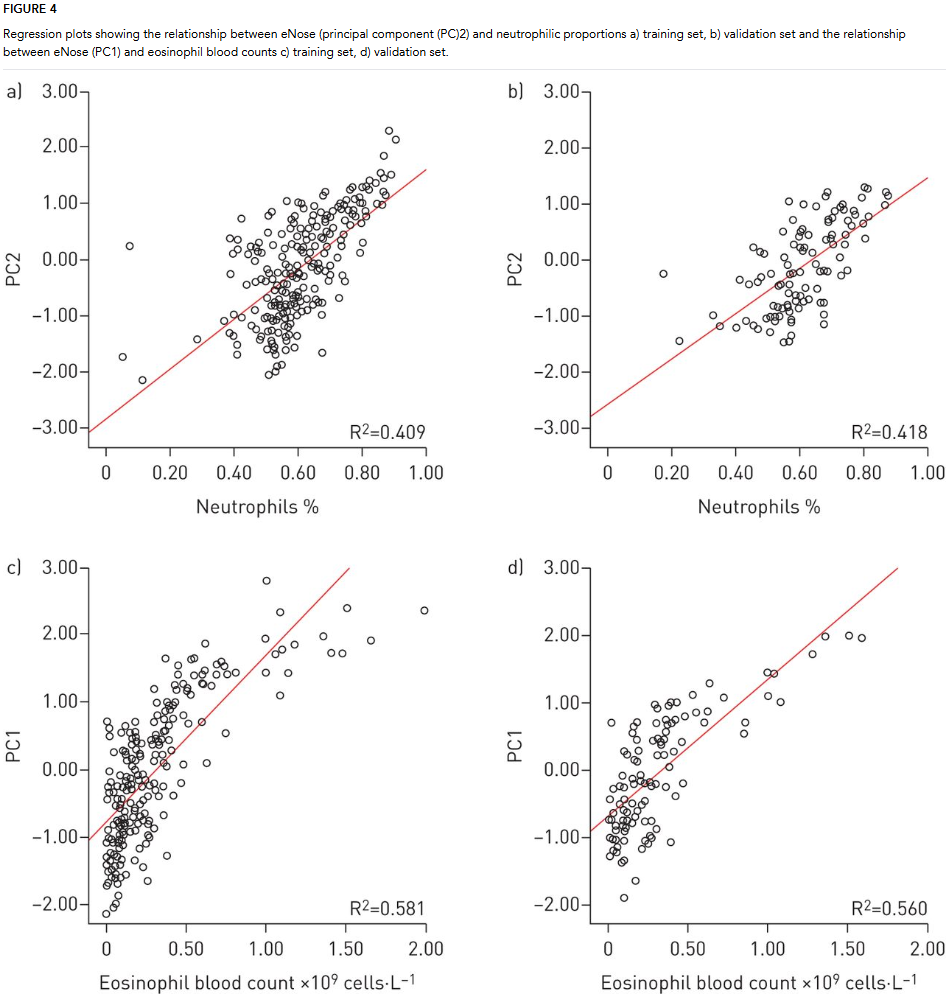
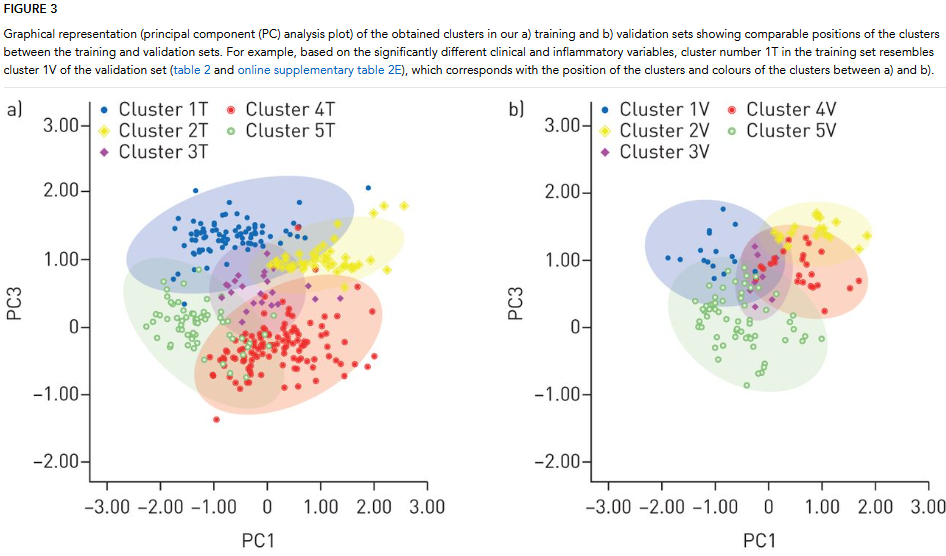
Data Analysis: Raw sensor data were processed using Matlab, focusing on peak signals and breath-hold ratios. A sample size calculation determined the need for 180 participants in the training set, with a smaller validation set included for additional reliability. Principal component analysis (PCA) was used to reduce data dimensionality. Four principal components explained over 91% of the variance. Hierarchical clustering identified significant clusters within the data, validated across both training and validation sets. Multiple linear regression was used to predict eosinophil and neutrophil counts based on eNose data. The model was validated, demonstrating strong predictive performance.
Results
The study analyzed exhaled breath profiles using eNose technology to identify clinical and inflammatory phenotypes in patients with asthma and COPD, without relying on traditional diagnostic labels. It included a training set of 206 asthma and 115 COPD patients, and a validation set of 114 patients. The baseline characteristics showed significant differences in age, lung function, and inflammatory markers between the groups. Using unsupervised cluster analysis, the researchers identified five distinct eNose-driven clusters in both the training and validation sets, highlighting different clinical and inflammatory traits, including eosinophilia, neutrophilia, atopy, and exacerbation rates.
The study found that the eNose could accurately predict systemic inflammatory markers like eosinophil and neutrophil counts using principal component analysis, with significant regression models confirmed in the validation set. Despite overlapping profiles between asthma and COPD patients, particularly those with comorbid conditions, the analysis identified distinct inflammatory phenotypes across the clusters. These findings suggest that eNose technology may serve as a valuable tool for phenotyping chronic airway diseases based on underlying inflammatory patterns rather than traditional diagnostic categories, supporting a more tailored approach to patient care.